Tidal marsh sparrowsEcology, Genetics, and Conservation Since 2011, we have been collecting demographic data from tidal marsh nesting birds on 4 long-term study sites in Maine, New Hampshire, and Massachusetts. We conduct mark-recapture banding and monitor nesting success of Saltmarsh and Nelson's sparrows to track long-term patterns in reproduction and survival. We are also investigating plasticity of nesting behaviors, offspring sex ratios, factors that influence paternity, and migratory movements with nanotags. Using genetic markers we have been investigating interspecific hybridization between Saltmarsh and Nelson's sparrows and differential fitness of pure species and hybrids. We are also working with collaborators to trial management actions, including floating habitat islands, to conserve these marsh-nesting sparrows, which are threatened by sea-level rise impacts on marsh habitats. Our tidal marsh bird research is part of a collaboration with the Saltmarsh Habitat and Avian Research Project, a group of academic, governmental, and non-profit collaborators focused on the conservation of tidal marsh birds. More information on this project can be found here. |
|
Ecological Genomics
The harsh abiotic environments of tidal marshes present adaptive challenges to vertebrates. We are using genomic tools of RAD Sequencing and whole genome sequencing in tidal marsh sparrows to investigate adaptive divergence across environmental gradients of the coastal-inland marsh ecotone, identify genes underlying traits conferring adaptation to tidal marshes, and address questions about parallel evolution and the genomic architecture of adaptation. We are working with researchers at the University of Maine in a collaborative NSF ESPCoR Track 2 Genomes-to-Phenomes project - the Genomic Ecology of Coastal Organisms (GECO). |
New England cottontail conservationMonitoring Occupancy & Abundance
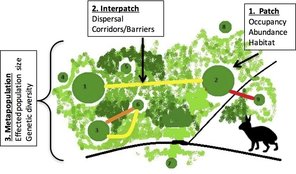
To aid conservation efforts for this species of concern, we work with natural resource managers to develop and implement monitoring efforts using noninvasive genetic sampling approaches. From fecal pellet surveys, we use species-diagnostic genetic assays to monitor the distribution and model occupancy patterns for New England cottontail. We recently developed a spatially explicit genetic mark-recapture abundance estimation framework, which we are beginning to implement on sites of management concern throughout the species range. Future work will model extinction/recolonization dynamics within a metapopulation context. Population & Conservation Genetics From the genetic data collected in fecal pellet surveys, we identify patterns of population structure and gene flow. Here as well, we take a metapopulation approach, and aim to identify the boundaries of interconnected, functioning metapopulations within the landscape. We monitor within population genetic diversity and work with managers to track the success of translocations and releases of cottontails from the captive breeding program. Landscape Genetics & Connectivity Using the genetic data in conjunction with GIS mapping and spatial statistics, we model cottontail dispersal and how it is influenced by features of the landscape. By implementing landscape genetic studies in multiple diverse landscapes across the species range, we are testing hypotheses about habitat fragmentation impacts and identifying critical dispersal corridors to guide restoration efforts. Learn More Our work with the New England cottontail is a component of a larger conservation initiative, which you can read about here. A general overview of our contributions to that initiative can be found here. |
Occupancy patterns of shrubland birdsExtensive habitat restoration efforts across the Northeast are focused on creating shrubland habitat for New England cottontail. Numerous other shrubland dependent species may benefit from these efforts, including shrubland birds. In this project, we are characterizing occupancy patterns of shrubland birds on sites occupied by and managed for New England cottontail. Our aim is to identify management practices and fine scale habitat characteristics that benefit particular suites of shrubland bird species. We also collaborate with UNH colleague Matt Tarr to further investigate these relationships in recent clear cuts and utility rights of way.
|
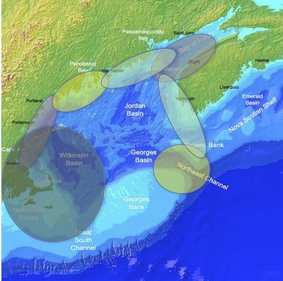
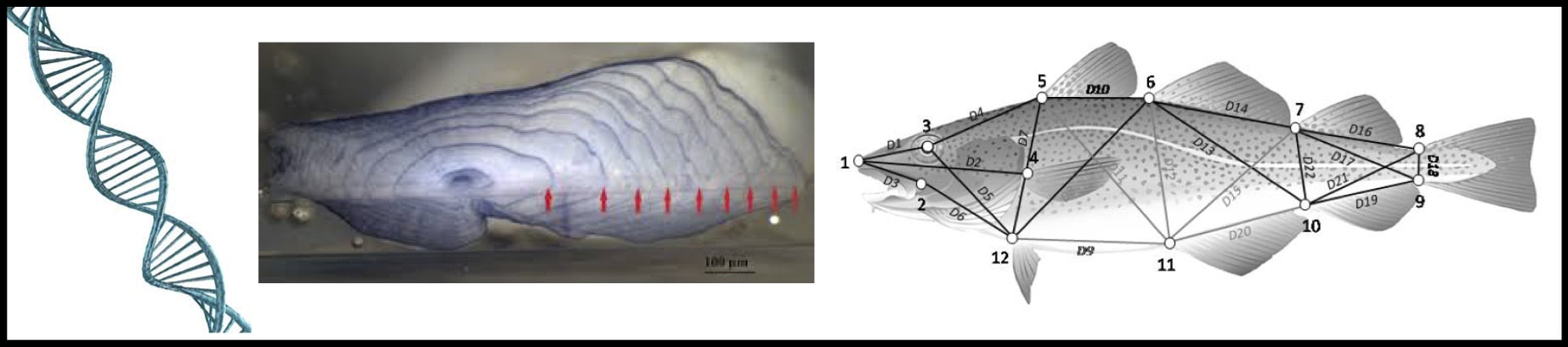
Stock identification of Atlantic cod
|
Striped Bass Population Structure & Strain Evaluation
|
In a NH Sea Grant- funded project we are working with David Belinsky (UNH) to evaluate the population origin of striped bass in a mixed fishery in coastal waters of northern and southern New England. We are using a ddRAD-Sequencing approach to develop a panel of high resolution markers for population assignment, which we will then apply to stripers caught in the mixed fishery in 3 different locations over 3 seasons. We will also use our results to inform aquaculture practices via population-of-origins-related differences in growth rates for stripers of different strains (river origins).
|
|